Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5(6):463–6.
Bodaghi A, Fattahi N, Ramazani A. Biomarkers. Promising and valuable tools towards diagnosis, prognosis and treatment of Covid-19 and other diseases. Heliyon. 2023;9(2):e13323.
Safa A, Bahroudi Z, Shoorei H, Majidpoor J, Abak A, Taheri M, et al. MiR-1: a comprehensive review of its role in normal development and diverse disorders. Biomed Pharmacother. 2020;132:110903.
Ghafouri-Fard S, Khoshbakht T, Hussen BM, Abdullah ST, Taheri M, Samadian M. A review on the role of mir-16-5p in the carcinogenesis. Cancer Cell Int. 2022;22(1):342.
Ghafouri-Fard S, Vafaee R, Shoorei H, Taheri M. Micrornas in gastric cancer: biomarkers and therapeutic targets. Gene. 2020;757:144937.
Hussen BM, Azimi T, Abak A, Hidayat HJ, Taheri M, Ghafouri-Fard S. Role of lncrna BANCR in human cancers: an updated review. Front Cell Dev Biol. 2021;9:689992.
Ghaforui-Fard S, Vafaee R, Taheri M. Taurine-upregulated gene 1: a functional long noncoding RNA in tumorigenesis. J Cell Physiol. 2019;234(10):17100–12.
Martinez-Dominguez MV, Zottel A, Šamec N, Jovčevska I, Dincer C, Kahlert UD, et al. Current technologies for RNA-directed liquid diagnostics. Cancers (Basel). 2021;13(20):13:20.
Martinez-Ledesma E, Verhaak RG, Treviño V. Identification of a multi-cancer gene expression biomarker for cancer clinical outcomes using a network-based algorithm. Sci Rep. 2015;5:11966.
Das S, Dey MK, Devireddy R, Gartia MR. Biomarkers in cancer detection, diagnosis, and prognosis. Sens. 2024; 24(1).
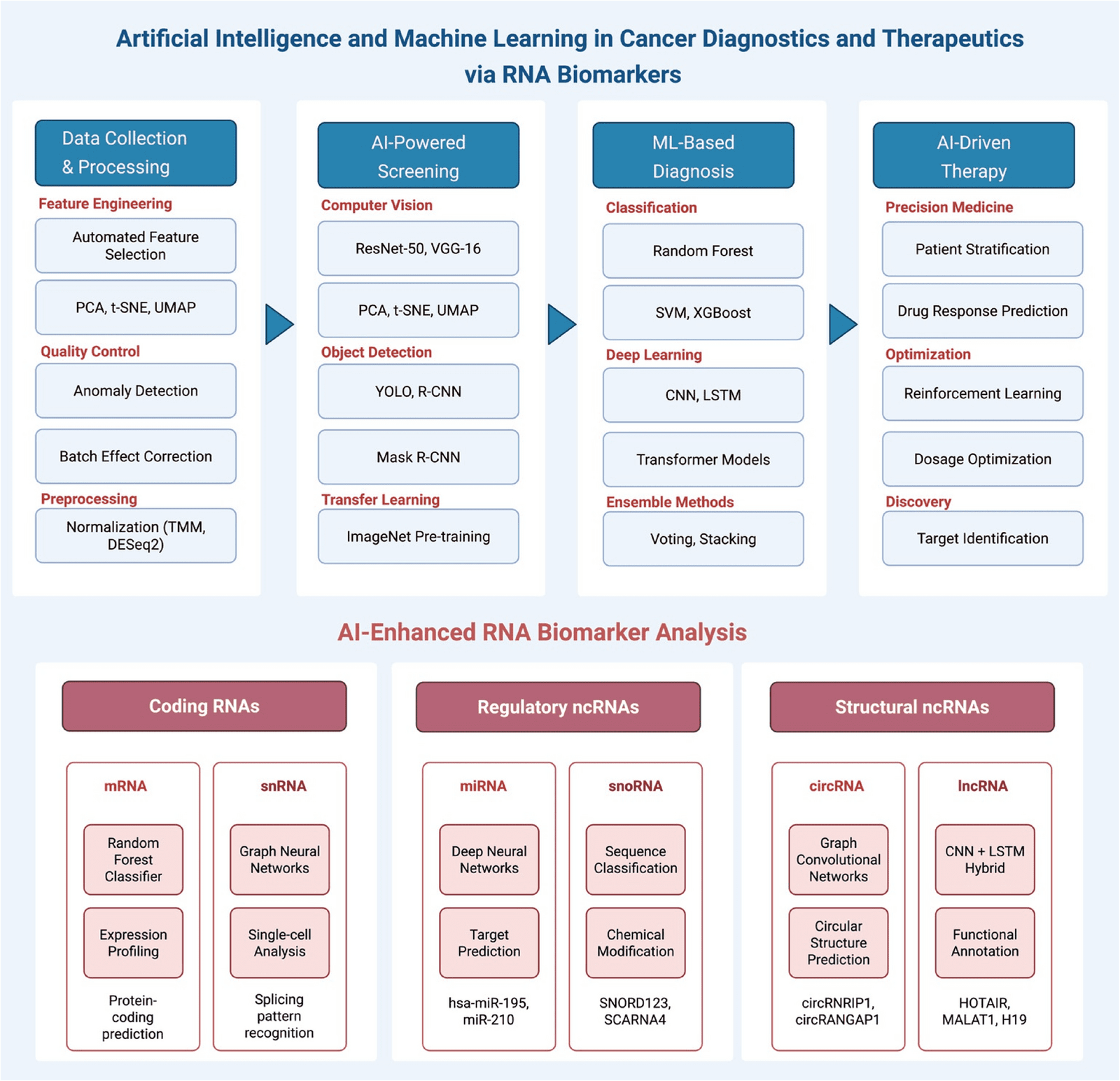
Quazi S. Artificial intelligence and machine learning in precision and genomic medicine. Med Oncol. 2022;39(8):120.
Santorsola M, Lescai F. The promise of explainable deep learning for omics data analysis: adding new discovery tools to AI. New Biotechnol. 2023;77:1–11.
Abdullah R, Masseh H, Salihi A, Faraj B. Unleashing the power of OpenAI in shaping the future of cancer research. BioMed Target Journal. 2023;1(1):2–11.
Aswathy R, Chalos VA, Suganya K, Sumathi S. Advancing miRNA cancer research through artificial intelligence: from biomarker discovery to therapeutic targeting. Med Oncol. 2024;42(1):30.
Shams A. Leveraging state-of-the-art AI algorithms in personalized oncology: from transcriptomics to treatment. Diagnostics. 2024;14(19):2174.
Wang J, Zeng Z, Li Z, Liu G, Zhang S, Luo C, et al. The clinical application of artificial intelligence in cancer precision treatment. J Transl Med. 2025;23(1):120.
Chen S-T, Hsiao Y-H, Huang Y-L, Kuo S-J, Tseng H-S, Wu H-K, et al. Comparative analysis of logistic regression, support vector machine and artificial neural network for the differential diagnosis of benign and malignant solid breast tumors by the use of three-dimensional power doppler imaging. Korean J Radiol. 2009;10(5):464–71.
Mahmood D, Aminfar S. Efficient machine learning and deep learning techniques for detection of breast cancer tumor. BioMed Target Journal. 2024;2:1–13.
Sofi PA, Zargar SM, Hamadani A, Shafi S, Zaffar A, Riyaz I, et al. Decoding life: genetics, bioinformatics, and artificial intelligence. A biologist s guide to artificial intelligence. Elsevier; 2024. pp. 47–66.
Wang T, Wang S, Li Z, Xie J, Jia Q, Hou J. Integrative machine learning model of RNA modifications predict prognosis and treatment response in patients with breast cancer. Cancer Cell Int. 2025;25:43.
Zhou X, Qu M, Tebon P, Jiang X, Wang C, Xue Y, et al. Screening cancer immunotherapy: when engineering approaches meet artificial intelligence. Adv Sci. 2020;7(19):2001447.
Bereczki Z, Benczik B, Balogh OM, Marton S, Puhl E, Pétervári M, et al. Mitigating off-target effects of small RNAs: conventional approaches, network theory and artificial intelligence. Br J Pharmacol. 2025;182(2):340–79.
Hu B, Yang YT, Huang Y, Zhu Y, Lu ZJ. POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 2017;45(D1):D104-14.
Lopez JP, Diallo A, Cruceanu C, Fiori LM, Laboissiere S, Guillet I, et al. Biomarker discovery: quantification of micrornas and other small non-coding RNAs using next generation sequencing. BMC Med Genomics. 2015;8:35.
Hu L, Xu Z, Hu B, Lu ZJ. COME: a robust coding potential calculation tool for LncRNA identification and characterization based on multiple features. Nucleic Acids Res. 2017;45(1):e2.
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–7.
Sarhadi VK, Armengol G. Molecular biomarkers in cancer. Biomolecules. 2022. https://doi.org/10.3390/biom12081021.
Hrašovec S, Glavač D. Micrornas as novel biomarkers in colorectal cancer. Front Genet. 2012;3:180.
Shah SNA, Parveen R. Differential gene expression analysis and machine learning identified structural, TFs, cytokine and glycoproteins, including SOX2, TOP2A, SPP1, COL1A1, and TIMP1 as potential drivers of lung cancer. Biomarkers. 2025;30(2):200–15.
Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–87.
Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers (Basel). 2019;11:8.
Norouzi-Barough L, Asgari Khosro Shahi A, Mohebzadeh F, Masoumi L, Haddadi MR, Shirian S. Early diagnosis of breast and ovarian cancers by body fluids circulating tumor-derived exosomes. Cancer Cell Int. 2020;20(1):187.
Spanos M, Gokulnath P, Chatterjee E, Li G, Varrias D, Das S. Expanding the horizon of EV-RNAs: LncRNAs in EVs as biomarkers for disease pathways. Extracellular Vesicle. 2023;2:100025.
Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Agarwal R, Kumar B, Jayadev M, Raghav D, Singh A. CoReCG: a comprehensive database of genes associated with colon-rectal cancer. Database (Oxford). 2016;2016.
Poos K, Smida J, Nathrath M, Maugg D, Baumhoer D, Neumann A et al. Structuring osteosarcoma knowledge: an osteosarcoma-gene association database based on literature mining and manual annotation. Database (Oxford). 2014;2014.
Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO. MIRUMIR: an online tool to test MicroRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell Death Differ. 2013;20(2):367.
Ainsztein AM, Brooks PJ, Dugan VG, Ganguly A, Guo M, Howcroft TK, et al. The NIH Extracellular RNA Communication Consortium. J Extracell Vesicles. 2015;4:27493.
Russo F, Di Bella S, Nigita G, Macca V, Laganà A, Giugno R, et al. MiRandola: extracellular circulating MicroRNAs database. PLoS ONE. 2012;7(10):e47786.
Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal. 2015;2015:657086.
Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. Exocarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40(Database issue):D1241-4.
Dlamini Z, Francies FZ, Hull R, Marima R. Artificial intelligence (AI) and big data in cancer and precision oncology. Comput Struct Biotechnol J. 2020;18:2300–11.
Pinto-Coelho L. How artificial intelligence is shaping medical imaging technology: a survey of innovations and applications. Bioengineering. 2023. https://doi.org/10.3390/bioengineering10121435.
Karalis VD. The integration of artificial intelligence into clinical practice. Appl Biosci. 2024;3(1):14–44.
Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17.
McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577(7788):89–94.
Kim HE, Kim HH, Han BK, Kim KH, Han K, Nam H, et al. Changes in cancer detection and false-positive recall in mammography using artificial intelligence: a retrospective, multireader study. Lancet Digit Health. 2020;2(3):e138–48.
Han SS, Park I, Eun Chang S, Lim W, Kim MS, Park GH, et al. Augmented intelligence dermatology: deep neural networks empower medical professionals in diagnosing skin cancer and predicting treatment options for 134 skin disorders. J Invest Dermatol. 2020;140(9):1753–61.
Undru TR, Uday U, Lakshmi JT, Kaliappan A, Mallamgunta S, Nikhat SS, et al. Integrating artificial intelligence for clinical and laboratory Diagnosis – a review. Maedica (Bucur). 2022;17(2):420–6.
Alowais SA, Alghamdi SS, Alsuhebany N, Alqahtani T, Alshaya AI, Almohareb SN, et al. Revolutionizing healthcare: the role of artificial intelligence in clinical practice. BMC Med Educ. 2023;23(1):689.
Peiffer-Smadja N, Dellière S, Rodriguez C, Birgand G, Lescure FX, Fourati S, et al. Machine learning in the clinical microbiology laboratory: has the time come for routine practice? Clin Microbiol Infect. 2020;26(10):1300–9.
Hull R, Mbele M, Makhafola T, Hicks C, Wang SM, Reis RM, et al. Cervical cancer in low and middle–income countries (review). Oncol Lett. 2020;20(3):2058–74.
Chuang L, Rainville N, Byrne M, Randall T, Schmeler K. Cervical cancer screening and treatment capacity: a survey of members of the African organisation for research and training in cancer (AORTIC). Gynecol Oncol Rep. 2021;38:100874.
Holmström O, Linder N, Lundin M, Moilanen H, Suutala A, Turkki R, et al. Quantification of estrogen receptor-Alpha expression in human breast carcinomas with a miniaturized, low-cost digital microscope: a comparison with a high-end whole slide-scanner. PLoS ONE. 2015;10(12):e0144688.
Salto-Tellez M, Maxwell P, Hamilton P. Artificial intelligence-the third revolution in pathology. Histopathology. 2019;74(3):372–6.
Lancellotti C, Cancian P, Savevski V, Kotha SRR, Fraggetta F, Graziano P, et al. Artificial intelligence & tissue biomarkers: advantages, risks and perspectives for pathology. Cells. 2021;10(4):787.
Kawka M, Dawidziuk A, Jiao LR, Gall TMH. Artificial intelligence in the detection, characterisation and prediction of hepatocellular carcinoma: a narrative review. Transl Gastroenterol Hepatol. 2022;7:41.
Gupta R, Kleinjans J, Caiment F. Identifying novel transcript biomarkers for hepatocellular carcinoma (HCC) using RNA-seq datasets and machine learning. BMC Cancer. 2021;21(1):962.
Zhou P, Li Z, Liu F, Kwon E, Hsieh TC, Ye S, et al. BAMBI integrates biostatistical and artificial intelligence methods to improve RNA biomarker discovery. Brief Bioinform. 2025. https://doi.org/10.1093/bib/bbaf073.
Wang T, Shao W, Huang Z, Tang H, Zhang J, Ding Z, et al. Mogonet integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nat Commun. 2021;12(1):3445.
Kurt F, Agaoglu M, Arga KY. Precision oncology: an ensembled machine learning approach to identify a candidate mRNA panel for stratification of patients with breast cancer. OMICS. 2022;26(9):504–11.
Turai PI, Herold Z, Nyirő G, Borka K, Micsik T, Tőke J, et al. Tissue miRNA combinations for the differential diagnosis of adrenocortical carcinoma and adenoma established by artificial intelligence. Cancers (Basel). 2022;14(4):895.
Zhao X, Dou J, Cao J, Wang Y, Gao Q, Zeng Q, et al. Uncovering the potential differentially expressed miRNAs as diagnostic biomarkers for hepatocellular carcinoma based on machine learning in the cancer genome atlas database. Oncol Rep. 2020;43(6):1771–84.
Sathipati SY, Tsai M-J, Shukla SK, Ho S-Y. Artificial intelligence-driven pan-cancer analysis reveals miRNA signatures for cancer stage prediction. Hum Genet Genom Adv. 2023. https://doi.org/10.1016/j.xhgg.2023.100190.
Hamidi F, Gilani N, Belaghi RA, Sarbakhsh P, Edgünlü T, Santaguida P. Exploration of potential miRNA biomarkers and prediction for ovarian cancer using artificial intelligence. Front Genet. 2021;12:724785.
Zhang X, Zhang H, Shen B, Sun X-F. Novel MicroRNA biomarkers for colorectal cancer early diagnosis and 5-fluorouracil chemotherapy resistance but not prognosis: a study from databases to AI-assisted verifications. Cancers. 2020;12(2):341.
Aghayousefi R, Hosseiniyan Khatibi SM, Zununi Vahed S, Bastami M, Pirmoradi S, Teshnehlab M. A diagnostic MiRNA panel to detect recurrence of ovarian cancer through artificial intelligence approaches. J Cancer Res Clin Oncol. 2023;149(1):325–41.
Li Q, Liu X, Gu J, Zhu J, Wei Z, Huang H. Screening lncrnas with diagnostic and prognostic value for human stomach adenocarcinoma based on machine learning and mrna-lncrna co‐expression network analysis. Mol Genet Genomic Med. 2020;8(11):e1512.
Liu Z, Guo C, Dang Q, Wang L, Liu L, Weng S et al. Integrative analysis from multi-center studies identities a consensus machine learning-derived LncRNA signature for stage II/III colorectal cancer. EBioMedicine. 2022;75.
Yu Y, Ren W, He Z, Chen Y, Tan Y, Mao L, et al. Machine learning radiomics of magnetic resonance imaging predicts recurrence-free survival after surgery and correlation of LncRNAs in patients with breast cancer: a multicenter cohort study. Breast Cancer Res. 2023;25(1):132.
Yang L, Cui Y, Liang L, Lin J. Significance of cuproptosis-related LncRNA signature in LUAD prognosis and immunotherapy: A machine learning approach. Thorac Cancer. 2023;14(16):1451–66.
Luo M, Lan F, Yang C, Ji T, Lou Y, Zhu Y, et al. Sensitive small extracellular vesicles associated circrnas analysis combined with machine learning for precision identification of gastric cancer. Chem Eng J. 2024;491:152094.
Pan X, Chen L, Feng K-Y, Hu X-H, Zhang Y-H, Kong X-Y, et al. Analysis of expression pattern of snornas in different cancer types with machine learning algorithms. Int J Mol Sci. 2019;20(9):2185.
Xing L, Zhang X, Zhang X, Tong D. Expression scoring of a small-nucleolar‐RNA signature identified by machine learning serves as a prognostic predictor for head and neck cancer. J Cell Physiol. 2020;235(11):8071–84.
Gholizadeh M, Mazlooman SR, Hadizadeh M, Drozdzik M, Eslami S. Detection of key mRNAs in liver tissue of hepatocellular carcinoma patients based on machine learning and bioinformatics analysis. MethodsX. 2023;10:102021.
Zhang Z, Liu ZP. Robust biomarker discovery for hepatocellular carcinoma from high-throughput data by multiple feature selection methods. BMC Med Genomics. 2021;14(Suppl 1):112.
Likhitrattanapisal S, Tipanee J, Janvilisri T. Meta-analysis of gene expression profiles identifies differential biomarkers for hepatocellular carcinoma and cholangiocarcinoma. Tumour Biol. 2016;37(9):12755–66.
Liao W, Xu Y, Pan M, Chen H. Serum micro-RNAs with mutation-targeted RNA modification: a potent cancer detection tool constructed using an optimized machine learning workflow. Sci Rep. 2024;14(1):9016.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. Microrna expression profiles classify human cancers. Nature. 2005;435(7043):834–8.
Shi W, Wartmann T, Accuffi S, Al-Madhi S, Perrakis A, Kahlert C, et al. Integrating a MicroRNA signature as a liquid biopsy-based tool for the early diagnosis and prediction of potential therapeutic targets in pancreatic cancer. Br J Cancer. 2024;130(1):125–34.
Yerukala Sathipati S, Tsai M-J, Shukla SK, Ho S-Y. Artificial intelligence-driven pan-cancer analysis reveals MiRNA signatures for cancer stage prediction. Hum Genet Genomics Adv. 2023. https://doi.org/10.1016/j.xhgg.2023.100190.
Metcalf GAD. Micrornas: circulating biomarkers for the early detection of imperceptible cancers via biosensor and machine-learning advances. Oncogene. 2024;43(28):2135–42.
Chi H, Chen H, Wang R, Zhang J, Jiang L, Zhang S, et al. Proposing new early detection indicators for pancreatic cancer: combining machine learning and neural networks for serum miRNA-based diagnostic model. Front Oncol. 2023;13:1244578.
Giuliano AE, Connolly JL, Edge SB, Mittendorf EA, Rugo HS, Solin LJ, et al. Breast cancer-major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(4):290–303.
Hussen BM, Hidayat HJ, Salihi A, Sabir DK, Taheri M, Ghafouri-Fard S. MicroRNA: a signature for cancer progression. Biomed Pharmacother. 2021;138:111528.
Begliarzade S, Beilerli A, Sufianov A, Tamrazov R, Kudriashov V, Ilyasova T, et al. Long non-coding RNAs as promising biomarkers and therapeutic targets in cervical cancer. Non-coding RNA Res. 2023;8(2):233–9.
Grosgeorges M, Picque Lasorsa L, Pastor B, Prévostel C, Crapez E, Sanchez C, et al. A straightforward method to quantify circulating mRNAs as biomarkers of colorectal cancer. Sci Rep. 2023;13(1):2739.
Wei L, Niraula D, Gates EDH, Fu J, Luo Y, Nyflot MJ, et al. Artificial intelligence (AI) and machine learning (ML) in precision oncology: a review on enhancing discoverability through multiomics integration. Br J Radiol. 2023. https://doi.org/10.1259/bjr.20230211.
Zahedi R, Ghamsari R, Argha A, Macphillamy C, Beheshti A, Alizadehsani R, et al. Deep learning in spatially resolved transcriptomics: a comprehensive technical view. Brief Bioinform. 2024. https://doi.org/10.1093/bib/bbae082.
Aldoseri A, Al-Khalifa KN, Hamouda AM. Re-thinking data strategy and integration for artificial intelligence: concepts, opportunities, and challenges. Appl Sci. 2023;13(12):7082.
Zhou LQ, Wu XL, Huang SY, Wu GG, Ye HR, Wei Q, et al. Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology. 2020;294(1):19–28.
Okugawa Y, Inoue Y, Tanaka K, Toiyama Y, Shimura T, Okigami M, et al. Loss of the metastasis suppressor gene KiSS1 is associated with lymph node metastasis and poor prognosis in human colorectal cancer. Oncol Rep. 2013;30(3):1449–54.
Zhang S, Zhang C, Du J, Zhang R, Yang S, Li B, et al. Prediction of lymph-node metastasis in cancers using differentially expressed mRNA and non-coding RNA signatures. Front Cell Dev Biol. 2021;9:605977.
Ji M, Wang W, Yan W, Chen D, Ding X, Wang A. Dysregulation of AKT1, a miR-138 target gene, is involved in the migration and invasion of tongue squamous cell carcinoma. J Oral Pathol Med. 2017;46(9):731–7.
Jin Y, Li Y, Wang X, Yang Y. Dysregulation of MiR-519d affects oral squamous cell carcinoma invasion and metastasis by targeting MMP3. J Cancer. 2019;10(12):2720–34.
Lee S, Cho M, Park B, Han K. Finding miRNA-RNA network biomarkers for predicting metastasis and prognosis in cancer. Int J Mol Sci. 2023. https://doi.org/10.3390/ijms24055052.
Li F, Yang M, Li Y, Zhang M, Wang W, Yuan D, et al. An improved clear cell renal cell carcinoma stage prediction model based on gene sets. BMC Bioinformatics. 2020;21(1):232.
Qureshi TA, Chen X, Xie Y, Murakami K, Sakatani T, Kita Y, et al. MRI/RNA-Seq-based radiogenomics and artificial intelligence for more accurate staging of muscle-invasive bladder cancer. Int J Mol Sci. 2023. https://doi.org/10.3390/ijms25010088.
Yerukala Sathipati S, Ho S-Y. Identifying a miRNA signature for predicting the stage of breast cancer. Sci Rep. 2018;8(1):16138.
Rodriguez-Luna H, Vargas HE, Byrne T, Rakela J. Artificial neural network and tissue genotyping of hepatocellular carcinoma in liver-transplant recipients: prediction of recurrence. Transplantation. 2005;79(12):1737–40.
Shen J, Qi L, Zou Z, Du J, Kong W, Zhao L, et al. Identification of a novel gene signature for the prediction of recurrence in HCC patients by machine learning of genome-wide databases. Sci Rep. 2020;10(1):4435.
Fu Y, Si A, Wei X, Lin X, Ma Y, Qiu H, et al. Combining a machine-learning derived 4-lncrna signature with AFP and TNM stages in predicting early recurrence of hepatocellular carcinoma. BMC Genomics. 2023;24(1):89.
Wang Z, Zhang N, Lv J, Ma C, Gu J, Du Y, et al. A five-gene signature for recurrence prediction of hepatocellular carcinoma patients. BioMed Res Int. 2020;2020(1):4037639.
Wang W, Wang L, Xie X, Yan Y, Li Y, Lu Q. A gene-based risk score model for predicting recurrence-free survival in patients with hepatocellular carcinoma. BMC Cancer. 2021;21(1):6.
Srividhya E, Niveditha VR, Nalini C, Sinduja K, Geeitha S. Integrating lncrna gene signature and risk score topredict recurrence cervical cancer using recurrent neural network. Meas Sensors. 2023;27:100782.
Levin Y, Talsania K, Tran B, Shetty J, Zhao Y, Mehta M. Optimization for sequencing and analysis of degraded FFPE-RNA samples. J Vis Exp. 2020(160).
Ottestad AL, Emdal EF, Grønberg BH, Halvorsen TO, Dai HY. Fragmentation assessment of FFPE DNA helps in evaluating NGS library complexity and interpretation of NGS results. Exp Mol Pathol. 2022;126:104771.
Joshi P, Dhar R, EpICC. A bayesian neural network model with uncertainty correction for a more accurate classification of cancer. Sci Rep. 2022;12(1):14628.
Erfanian N, Heydari AA, Feriz AM, Iañez P, Derakhshani A, Ghasemigol M, et al. Deep learning applications in single-cell genomics and transcriptomics data analysis. Biomed Pharmacother. 2023;165:115077.
Raza K. Preprocessing and Quality Control. 2024. p. 17-30.
Boland MR, Karczewski KJ, Tatonetti NP. Ten simple rules to enable multi-site collaborations through data sharing. PLoS Comput Biol. 2017;13(1):e1005278.
Zurek B, Ellwanger K, Vissers LELM, Schüle R, Synofzik M, Töpf A, et al. Solve-RD: systematic pan-European data sharing and collaborative analysis to solve rare diseases. Eur J Hum Genet. 2021;29(9):1325–31.
Ramzan F, Sartori C, Consoli S, Reforgiato Recupero D. Generative adversarial networks for synthetic data generation in finance: evaluating statistical similarities and quality assessment. AI. 2024;5(2):667–85.
Nirmal AJ, Maliga Z, Vallius T, Quattrochi B, Chen AA, Jacobson CA, et al. The spatial landscape of progression and immunoediting in primary melanoma at single-cell resolution. Cancer Discov. 2022;12(6):1518–41.
Frasca M, La Torre D, Repetto M, De Nicolò V, Pravettoni G, Cutica I. Artificial intelligence applications to genomic data in cancer research: a review of recent trends and emerging areas. Discover Analytics. 2024;2(1):10.
Alum EU. Ai-driven biomarker discovery: enhancing precision in cancer diagnosis and prognosis. Discover Oncol. 2025;16(1):313.
de Gonzalo-Calvo D, Karaduzovic-Hadziabdic K, Dalgaard LT, Dieterich C, Perez-Pons M, Hatzigeorgiou A, et al. Machine learning for catalysing the integration of noncoding RNA in research and clinical practice. EBioMedicine. 2024;106:105247.
Asim MN, Ibrahim MA, Imran Malik M, Dengel A, Ahmed S. Advances in computational methodologies for classification and Sub-Cellular locality prediction of Non-Coding RNAs. Int J Mol Sci. 2021;22(16).
Agajanian S, Oluyemi O, Verkhivker GM. Integration of random forest classifiers and deep convolutional neural networks for classification and biomolecular modeling of cancer driver mutations. Front Mol Biosci. 2019;6:44.
Yousef M, Goy G, Mitra R, Eischen CM, Jabeer A, Bakir-Gungor B. Mircorrnet: machine learning-based integration of miRNA and mRNA expression profiles, combined with feature grouping and ranking. PeerJ. 2021;9:e11458.
Dai C, Zeng X, Zhang X, Liu Z, Cheng S. Machine learning-based integration develops a mitophagy-related lncRNA signature for predicting the progression of prostate cancer: a bioinformatic analysis. Discover Oncol. 2024;15(1):316.
Xu J, Yang P, Xue S, Sharma B, Sanchez-Martin M, Wang F, et al. Translating cancer genomics into precision medicine with artificial intelligence: applications, challenges and future perspectives. Hum Genet. 2019;138(2):109–24.
Athanasopoulou K, Michalopoulou VI, Scorilas A, Adamopoulos PG. Integrating artificial intelligence in next-generation sequencing: advances, challenges, and future directions. Curr Issues Mol Biol. 2025;47(6):470.
Singh RP, Hom GL, Abramoff MD, Campbell JP, Chiang MF. Current challenges and barriers to real-world artificial intelligence adoption for the healthcare system, provider, and the patient. Transl Vis Sci Technol. 2020;9(2):45.
Goktas P, Grzybowski A. Shaping the future of healthcare: ethical clinical challenges and pathways to trustworthy AI. J Clin Med. 2025. https://doi.org/10.3390/jcm14051605.
Hanna MG, Pantanowitz L, Jackson B, Palmer O, Visweswaran S, Pantanowitz J, et al. Ethical and bias considerations in artificial intelligence/machine learning. Mod Pathol. 2025;38(3):100686.
Gupta S, Kumar S, Chang K, Lu C, Singh P, Kalpathy-Cramer J. Collaborative privacy-preserving approaches for distributed deep learning using multi-institutional data. Radiographics. 2023;43(4):e220107.